A Plant Cell paper is online now!
Topic - AS of RLI1 modulates plant Pi signaling and growth
Abstract
Phosphate (Pi) limitation represents a primary constraint on crop production. To better cope with Pi deficiency stress, plants have evolved multiple adaptive mechanisms for phosphorus acquisition and utilization, including the alteration of growth and the activation of Pi starvation signaling. However, how these strategies are coordinated remains largely unknown. Here, we found that the alternative splicing (AS) of REGULATOR OF LEAF INCLINATION 1 (RLI1) in rice (Oryza sativa) produces two protein isoforms: RLI1a, containing MYB DNA binding domain; and RLI1b, containing both MYB and coiled-coil (CC) domains. The absence of a CC domain in RLI1a enables it to activate broader target genes than RLI1b. RLI1a, but not RLI1b, regulates both brassinolide (BL) biosynthesis and signaling by directly activating BL-biosynthesis and signaling genes. Both RLI1a and RLI1b modulate Pi starvation signaling. RLI1 and PHOSPHATE STARVATION RESPONSE 2 function redundantly to regulate Pi starvation signaling and growth in response to Pi deficiency. Furthermore, the AS of RLI1-related genes to produce two isoforms for growth and Pi signaling is widely present in both dicots and monocots. Together, these findings indicate that the AS of RLI1 is an important and functionally conserved strategy to orchestrate Pi starvation signaling and growth to help plants adapt to Pi-limitation stress.
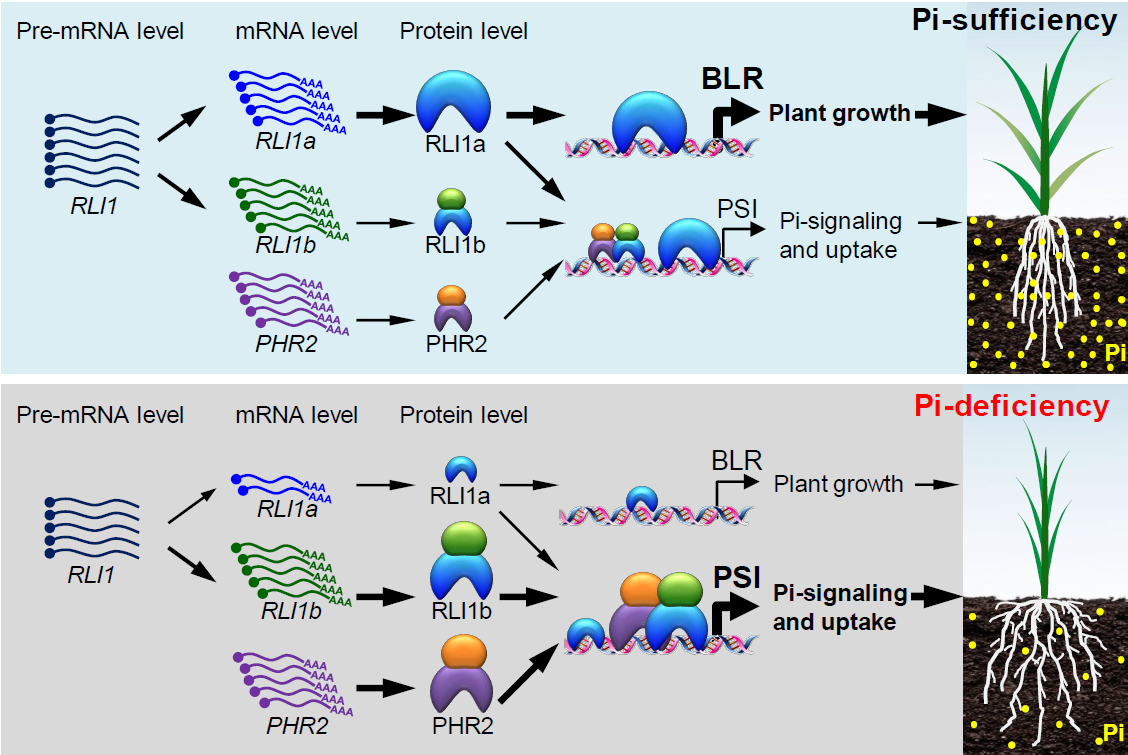
Working model of the role of RLI1 in regulating plant growth and Pi signaling to help plants adapt to Pi-deficiency
The thickness of the lines represents the degree of effects. The different sizes of protein diagrams represent protein levels. RLI1a targets downstream genes as a monomer. RLI1b and PHR2 target downstream genes as a dimer. A detailed description of the model can be found in the “Discussion.”
Link: https://doi.org/10.1093/plcell/koac161
News (In English)
Wechat promotion (In Chinese)